research
I am an ecologist and evolutionary biologist focused on problems in molecular ecology, distributional ecology, and biodiversity science. For the most up-to-date information on my research, please visit the Bagley Lab website as well as our updated Research page.
What follows is a listing of my research interests and projects primarily during my postdoc period (2015–2020). Click on the gray navigation bar above to quickly access research topics below.
phylogenomics & species delimitation
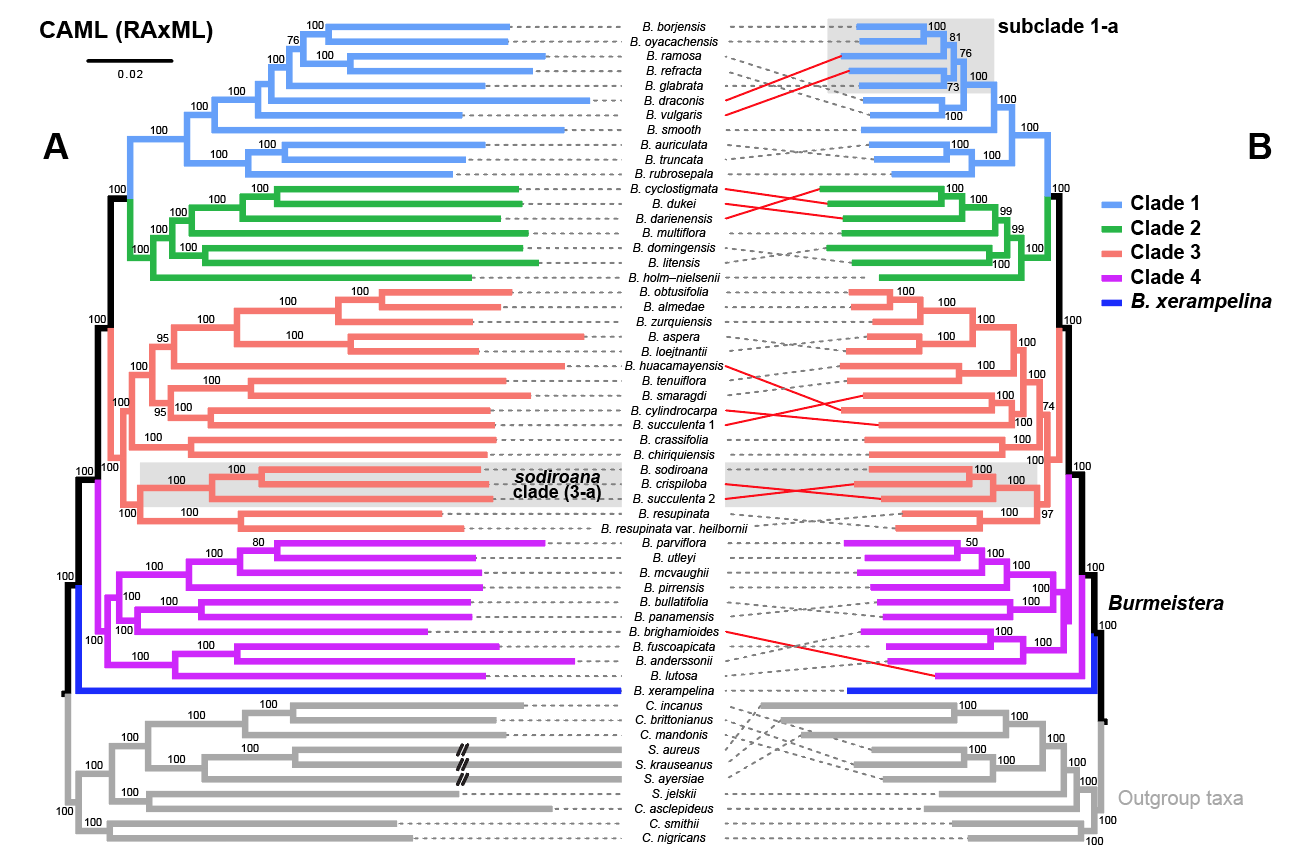
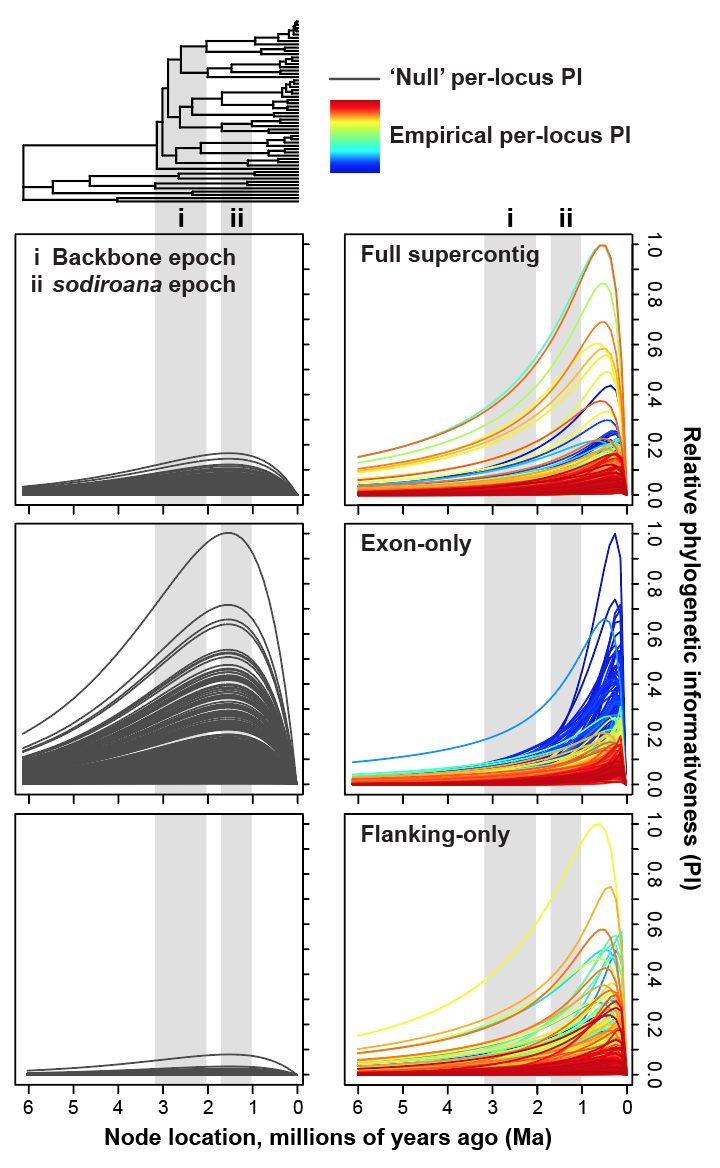
My research in systematics and taxonomy primarily focuses on discovering and determining evolutionary relationships among species, and clarifying species limits, through analyses of DNA sequence data and morphological characters. Hypotheses of phylogenetic relationships are reconstructed based on broad taxon and character sampling using maximum-likelihood and Bayesian inference analyses, relaxed molecular clocks, clock-partitioning schemes, and tip- and fossil-calibrated models (e.g. fossilized birth-death process). Our work also seeks to describe new biodiversity to improve biodiversity accounting. This work focuses on understanding diversification and the evolution of adaptive traits (e.g. reproductive mode) in monophyletic clades of temperate and Neotropical plants (bellflowers, pines), freshwater fishes, and squamate reptiles. Recent papers focus on
- –Phylogenomics of recent, rapid angiosperm radiations with Andean-centered distributions, using Burmeistera (Campanulaceae) as a case study
- –Integrating molecules, morphology, and fossils to infer relationships among North American 'sucker' fishes (Catostomidae)
- –Integrative taxonomy and species delimitation in Cerrado lizards.
phylogeography of freshwater & terrestrial ecosystems
|
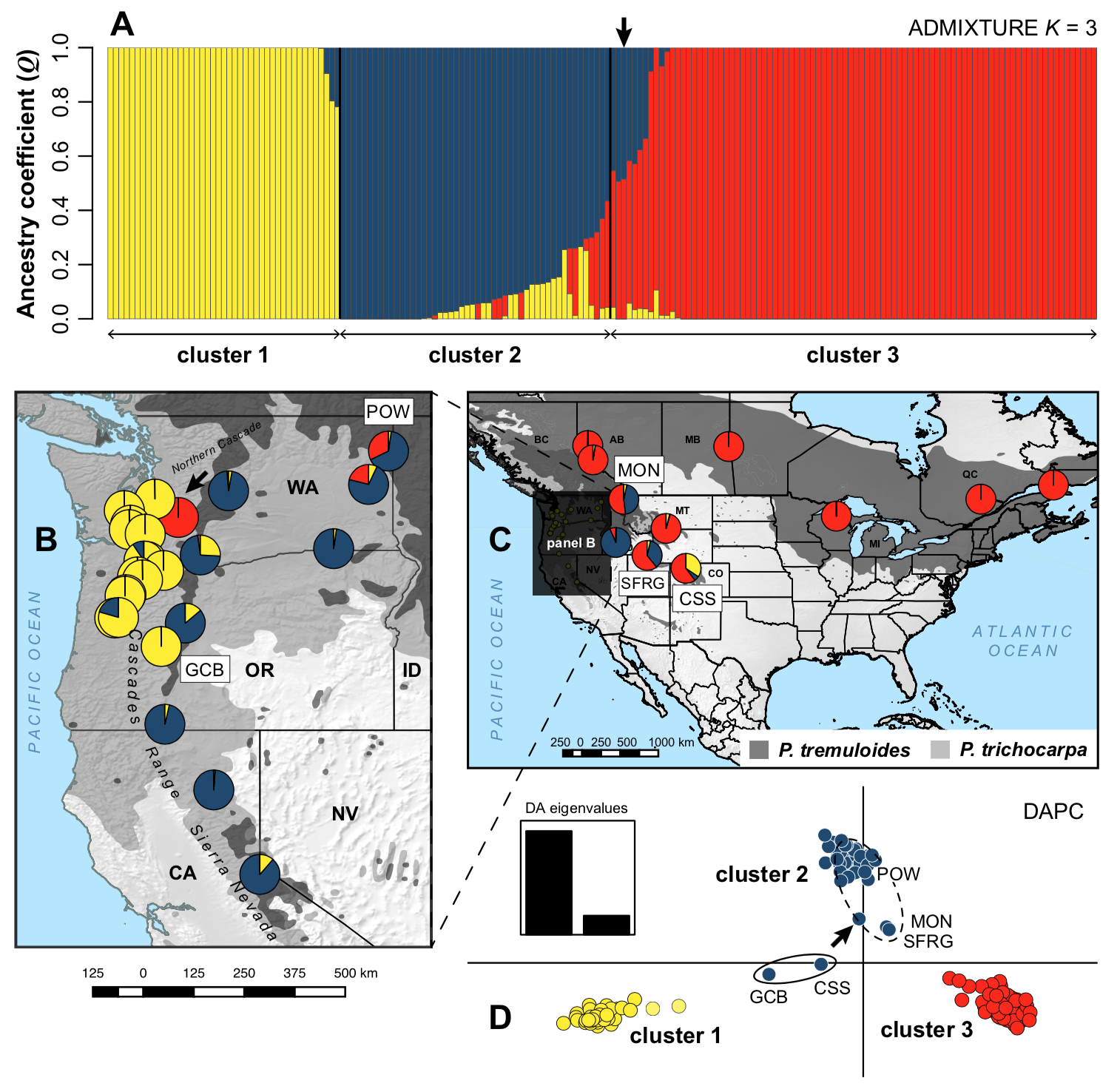
Phylogeography, the study of the geographical distributions of genetic lineages within and among closely related species, has assumed a central role in molecular ecology for inferring the historical and ecological processes influencing present-day biodiversity patterns. Along with collaborators from the U.S., Brazil, and Mexico, I have been involved in a number of projects using single-species and comparative (multispecies) phylogeography approaches to improve our understanding of the diversification, stability, and assembly of North American and Neotropical biotic communities. This work currently focuses on three intriguing systems that present different contexts for understanding evolutionary processes: 1) North American forest trees, and freshwater fish assemblages of the 2) Mesoamerica (Central America) and 3) Brazilian Cerrado biodiversity 'hotspots'.
Here is a more in-depth look at my prior research in phylogeography.
genomics of speciation & local adaptation, with implications for climate change
|
|
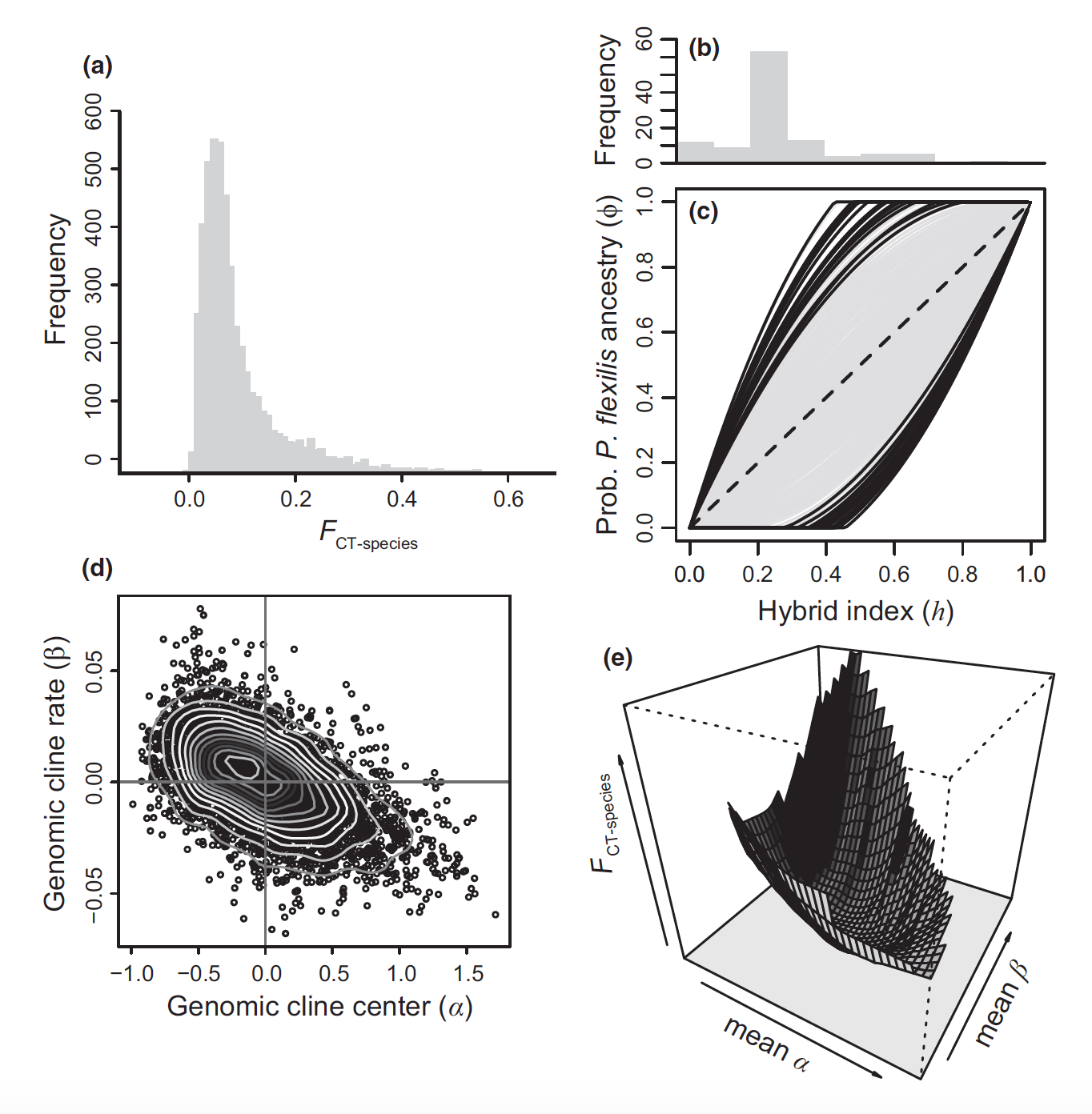
Understanding how barriers to gene flow arise, and new species boundaries are formed and maintained, is a central goal of evolutionary biology. Yet, the study of 'speciation', presents numerous challenges and requires integrative perspectives taking into account geographical factors (biogeography) and range dynamics, niche evolution, reproductive isolation, as well as the evolution of neutral and adaptive genetic variation in the context of demographic history. I have become interested in the ways in which plant versus animal systems can reveal different aspects of the speciation process.
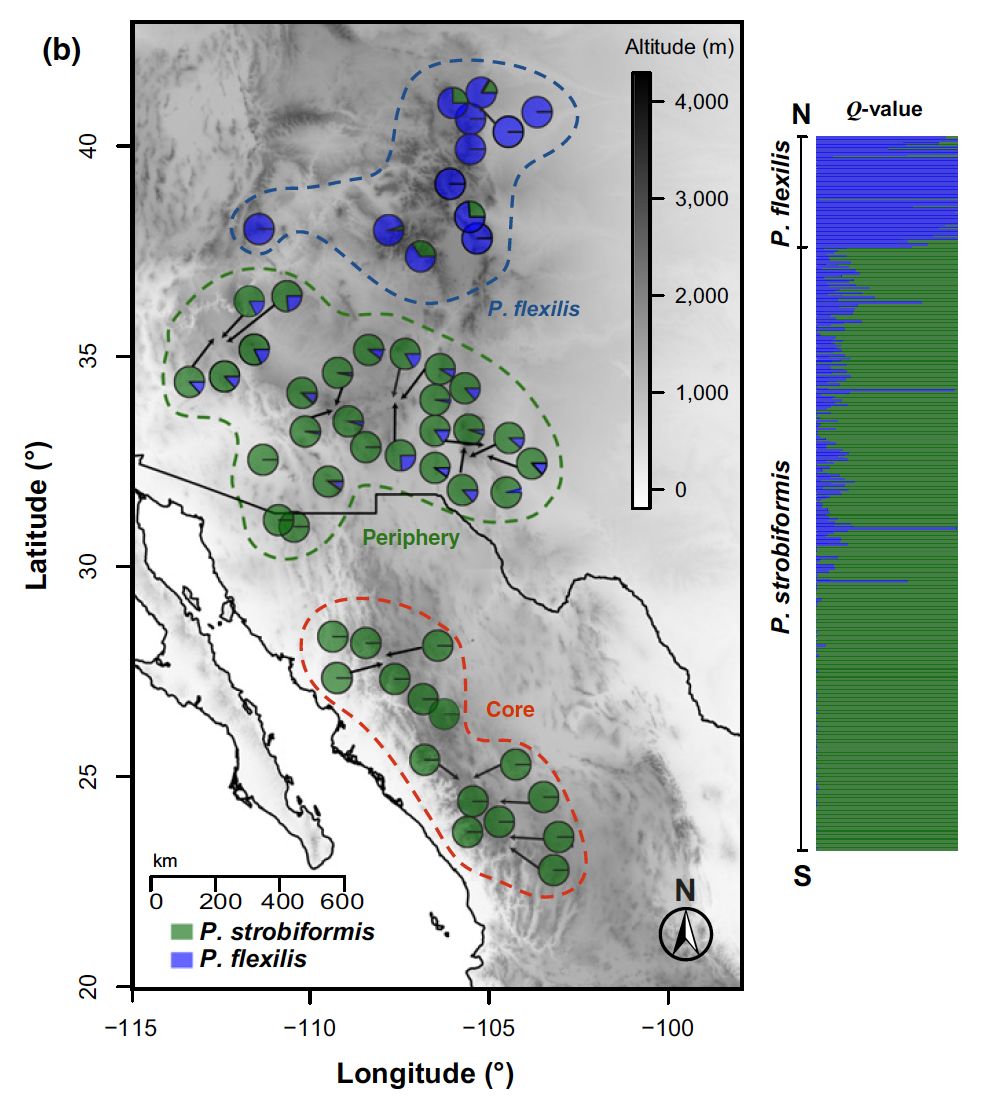
Our current projects on southwestern white pine (Pinus strobiformis) highlight the role of hybridization and extrinsic factors during ecological speciation. We are also using genome scan and environmental association analyses to understand local adaptation to climatic gradients and challenging environments (high elevation) in this system, with important implications for understanding tree responses to climate change. By contrast, some Neotropical fish lineages that I have studied (e.g. livebearers; Poeciliidae) are more predisposed to speciation through intrinsic pre- or postmating isolation, and exhibit presumed niche conservatism but a high degree of phenotypic variation. I am eager to develop future projects expanding my speciation and adaptation work along these two avenues of research.
code & bioinformatics pipelines
Check my GitHub and software pages to see what I've been coding lately. However, the main software package that I am currently developing is PIrANHA v0.4a4, a useful repository of pipelines and wrapper functions for automating tasks in phylogenetics and phylogeography.
What follows is a slightly dated summary of projects I am involved in, as recently as 2017–2018, which I hope to find time to update soon.
PhD dissertation research
[coming soon.]